DNA/RNA Mismatch
To generate models, user has to click either DNA or RNA button ( present at the bottom part of this page) then choose 'Ideal model' or 'Sequence specific model' or intramolecular DNA/RNA hairpin option and there insert the input sequences, with/without mismatch(es), in given places.
To built 'Sequence specific Model', user should choose the option of either NMR or X-RAY.
In addition, user can also generate intramolecular DNA/RNA duplexes (hairpin structures) for loop sequences by selecting 'DNA Hairpin' or 'RNA Hairpin' button in the given options.
The orientation of the input sequences will be considered as mentioned in the left side of the box.
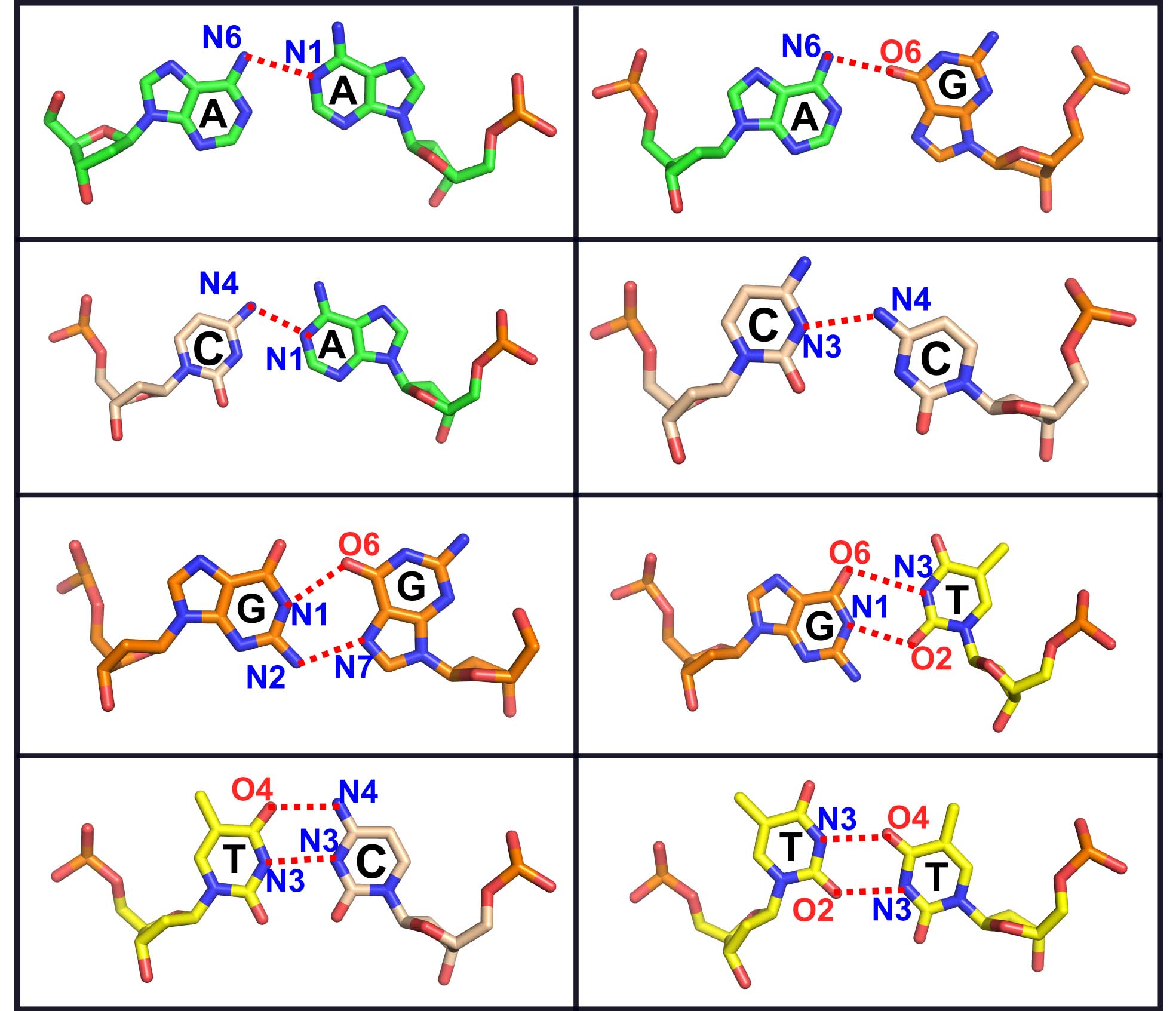
Click 'Submit' to generate the DNA/RNA molecule & information about the mismatch position(s).In the result page, mismatch is shown in different color in JSmol viewer.
3D-NuS displays the type of mismatch(es) and it's/their positions in 'Result' page. You can also view the generated 3D structure in the JSmol interactive viewer or can 'download' and visualize using other molecular visualizers. The hydrogen bonding schemes for DNA duplex is shown in Figure 1. For RNA, replace Thymine with Uracil.